The author is an associate professor of dairy cattle genetics at Penn State University.
One of the keys for a breed to remain economically viable is the ability to make genetic progress, and small populations are at a large disadvantage in this respect. Genomic selection may create an even larger economic disparity between large and small breeds as genetic progress accelerates in breeds that can implement robust genomic evaluations.
Small populations limit our ability to generate progeny tests in two important respects. First, there is a limit to the number of bulls that can be daughter-proven if there is a small population of cows to breed. We're not likely to find a sufficient number of exceptional bulls to improve a breed's genetic merit if we can't generate daughter-proven bulls. Second, bulls that are proven tend to have relatively few daughters. This means his daughter proof will be less accurate, particularly for lower heritability traits such as DPR (daughter pregnancy rate).
The challenges smaller breeds face when it comes to progeny tests carry through to our ability to conduct genomic evaluations. Accurate genomic predictions rely on having a robust population of animals that can be used to estimate the effect of DNA markers. We call this a predictor or reference population. As one would expect, a small predictor population with inaccurate genetic evaluations leads to lousy genomic predictions.
The focus of our effort to build our genomic prediction system has, to this point, largely centered on developing a robust bull predictor population. Bulls are considered more critical because they have highly accurate PTA (predicted transmitting ability) data when compared to cows. Sire genetic evaluations are based on the performance of many daughters spread across a large number of herds, whereas cow PTA depends on her own record and a handful of daughters, most often all from a single herd. There are some countries that do not include cows at all in their predictor population because they introduce a variety of challenges that can bias genomic PTA. That is not the situation in the United States as USDA scientists have developed methods that allow cow information to be successfully included.
Bulls trump cows
USDA scientists recently evaluated how much including cow information adds to reliability for Holstein genomic PTA. Their evaluation suggested that it adds less than 1 percent to the reliability of most traits when compared to what is achieved when evaluating only bulls.
I tend to think that may undervalue the influence of adding cows because a wider range of pedigrees is represented when compared to relying solely on daughter-proven bulls. Nevertheless, it is clear that using bull data has a stronger influence than cows for breeds with a large bull population.
While the concept of having a larger bull predictor population is sound, what should we do if that is simply not possible because of a small breed population? Recent research (Journal of Dairy Science, 97:58225832) suggests that genotyping a large proportion of the cows that are on DHI test has a greater importance for smaller breeds.
The authors compared predictor populations that considered genomically testing zero, 2,000 or 4,000 cows per year. Adding cows to the reference population improved genomic predictions considerably. The 4,000-cow threshold resulted in the highest reliabilities, but the gain when compared to testing 2,000 cows per year was limited.
In addition to improving reliability, testing cows will help to identify a larger variety of elite females to produce the next generation of young bulls. Genotyping a large number of cows also reduced the annual rate of inbreeding, which is a current challenge for our smaller populations.
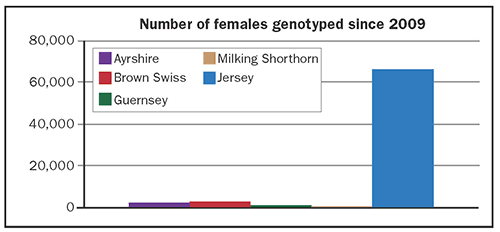
The chart shows the number of cows from each breed that have been genotyped with the exception of Holstein. I considered not including Jerseys in the chart, but left them in because it helps to demonstrate how far the other breeds are lagging in terms of the number of cows that have been genotyped. Fewer than 3,000 females of any of the smaller breeds have been genotyped since 2009, but Jerseys have over 66,000. By comparison, nearly 500,000 Holstein females have been genotyped.
There are around 2,000 Ayrshires and Guernseys born annually that contribute to national genetic evaluations and approximately 6,000 Brown Swiss. The study referenced above was for a slightly larger population and it would be difficult to reach the threshold of 2,000 cows genotyped per year, but there does appear to be an opportunity to test many more females.
Not represented in this chart is the sizable population of Brown Swiss bulls genotyped due to international collaboration. However, it is important to note that a static reference population will lose its value rapidly and that it must reflect current Brown Swiss genotypes. It may be difficult to grow the Brown Swiss bull reference population in future years and testing more females will increase in importance.
Raising the proportion of females that are genomically tested would provide a boost to the long-term development of the breeding programs of smaller breeds. There are challenges, including the costs associated with genomic testing.
In order for genomic testing to be most effective, we need breeders to test both their good cows and their poorer cows. It is hard from a producer perspective to justify spending money to test poorer cows. The cost of testing is especially difficult to consider for those with breeds, such as Guernseys, that don't have genomic PTA data available. Yet, until that initial investment in genetic testing is made, there will be no genomic evaluations.
A future must
Our smaller breeds have significant limitations when it comes to developing and maintaining a robust progeny test program. Genomic testing can help overcome this limitation to a degree, but I'm not sure we're realizing that potential. There are cost considerations to be sure, but more aggressive testing of cows that have their own performance records will help us take advantage of the opportunity to accelerate genetic gain.
This article appears on page 49 of the January 25, 2015 issue of Hoard's Dairyman.
Return to the Hoard's Dairyman feature page.
One of the keys for a breed to remain economically viable is the ability to make genetic progress, and small populations are at a large disadvantage in this respect. Genomic selection may create an even larger economic disparity between large and small breeds as genetic progress accelerates in breeds that can implement robust genomic evaluations.
Small populations limit our ability to generate progeny tests in two important respects. First, there is a limit to the number of bulls that can be daughter-proven if there is a small population of cows to breed. We're not likely to find a sufficient number of exceptional bulls to improve a breed's genetic merit if we can't generate daughter-proven bulls. Second, bulls that are proven tend to have relatively few daughters. This means his daughter proof will be less accurate, particularly for lower heritability traits such as DPR (daughter pregnancy rate).
The challenges smaller breeds face when it comes to progeny tests carry through to our ability to conduct genomic evaluations. Accurate genomic predictions rely on having a robust population of animals that can be used to estimate the effect of DNA markers. We call this a predictor or reference population. As one would expect, a small predictor population with inaccurate genetic evaluations leads to lousy genomic predictions.
The focus of our effort to build our genomic prediction system has, to this point, largely centered on developing a robust bull predictor population. Bulls are considered more critical because they have highly accurate PTA (predicted transmitting ability) data when compared to cows. Sire genetic evaluations are based on the performance of many daughters spread across a large number of herds, whereas cow PTA depends on her own record and a handful of daughters, most often all from a single herd. There are some countries that do not include cows at all in their predictor population because they introduce a variety of challenges that can bias genomic PTA. That is not the situation in the United States as USDA scientists have developed methods that allow cow information to be successfully included.
Bulls trump cows
USDA scientists recently evaluated how much including cow information adds to reliability for Holstein genomic PTA. Their evaluation suggested that it adds less than 1 percent to the reliability of most traits when compared to what is achieved when evaluating only bulls.
I tend to think that may undervalue the influence of adding cows because a wider range of pedigrees is represented when compared to relying solely on daughter-proven bulls. Nevertheless, it is clear that using bull data has a stronger influence than cows for breeds with a large bull population.
While the concept of having a larger bull predictor population is sound, what should we do if that is simply not possible because of a small breed population? Recent research (Journal of Dairy Science, 97:58225832) suggests that genotyping a large proportion of the cows that are on DHI test has a greater importance for smaller breeds.
The authors compared predictor populations that considered genomically testing zero, 2,000 or 4,000 cows per year. Adding cows to the reference population improved genomic predictions considerably. The 4,000-cow threshold resulted in the highest reliabilities, but the gain when compared to testing 2,000 cows per year was limited.
In addition to improving reliability, testing cows will help to identify a larger variety of elite females to produce the next generation of young bulls. Genotyping a large number of cows also reduced the annual rate of inbreeding, which is a current challenge for our smaller populations.

The chart shows the number of cows from each breed that have been genotyped with the exception of Holstein. I considered not including Jerseys in the chart, but left them in because it helps to demonstrate how far the other breeds are lagging in terms of the number of cows that have been genotyped. Fewer than 3,000 females of any of the smaller breeds have been genotyped since 2009, but Jerseys have over 66,000. By comparison, nearly 500,000 Holstein females have been genotyped.
There are around 2,000 Ayrshires and Guernseys born annually that contribute to national genetic evaluations and approximately 6,000 Brown Swiss. The study referenced above was for a slightly larger population and it would be difficult to reach the threshold of 2,000 cows genotyped per year, but there does appear to be an opportunity to test many more females.
Not represented in this chart is the sizable population of Brown Swiss bulls genotyped due to international collaboration. However, it is important to note that a static reference population will lose its value rapidly and that it must reflect current Brown Swiss genotypes. It may be difficult to grow the Brown Swiss bull reference population in future years and testing more females will increase in importance.
Raising the proportion of females that are genomically tested would provide a boost to the long-term development of the breeding programs of smaller breeds. There are challenges, including the costs associated with genomic testing.
In order for genomic testing to be most effective, we need breeders to test both their good cows and their poorer cows. It is hard from a producer perspective to justify spending money to test poorer cows. The cost of testing is especially difficult to consider for those with breeds, such as Guernseys, that don't have genomic PTA data available. Yet, until that initial investment in genetic testing is made, there will be no genomic evaluations.
A future must
Our smaller breeds have significant limitations when it comes to developing and maintaining a robust progeny test program. Genomic testing can help overcome this limitation to a degree, but I'm not sure we're realizing that potential. There are cost considerations to be sure, but more aggressive testing of cows that have their own performance records will help us take advantage of the opportunity to accelerate genetic gain.








